Deep Learning-based Cancer Biomarker Discovery with Graph Neural Networks using Attention-based Knowledge Graph Embeddings and Protein-Protein Interactions.
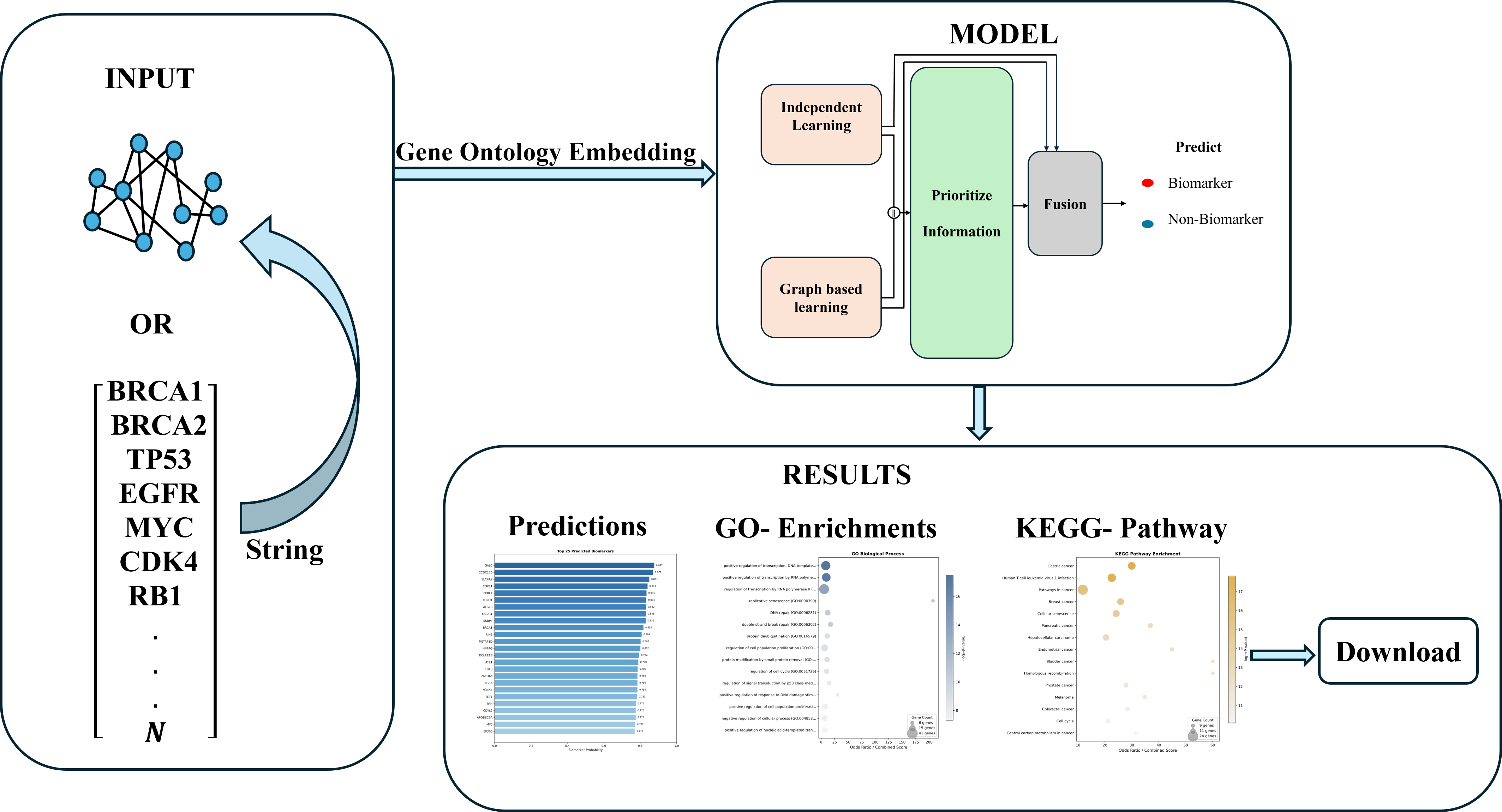
Overview
This tool integrates multiple data sources to identify potential cancer biomarkers using protein-protein interaction networks, Gene Ontology annotations, and knowledge graph embeddings. The prediction model uses Deep learning architecture trained on cancer-specific datasets.
Supported Cancer Types
Breast Cancer
Glioblastoma
Lung Cancer
Analysis Pipeline
- Input: Gene list or pre-constructed PPI network
- PPI Construction: Build network from STRING database
- Embedding Generation: Generate GO term embeddings using fine-tuned BioBERT
- Feature Assembly: Concatenate GO and GeoKG embeddings
- Prediction: Deep learning model based classification
- Enrichment Analysis: GO and KEGG pathway analysis
Global Usage
Researchers using GO-DMBC worldwide